Abstract
Seeds from wheat were tested for their aptitude to be transformed at early stages of germination by Agrobacterium tumefaciens, which promotes singular integration patterns, entailing lower mutational consequences for the transgenic plant and limited transgene silencing. Seeds were processed at different stages of germination by a cassette containing the GUS reporter gene under the control of a high molecular weight glutenin subunit promoter. Transgene integration and expression into the host organism were followed by PCR and by histological staining. Results showed that the cereals used could be effectively transformed by this method and that the foreign gene behaviour in the progenies was similar to that reported from the standard method, which uses explants and tissue culture. This simplified protocol is easy to use and allows savings in time and reagents. Moreover, it avoids the complex stages of tissue culture and plant regeneration. Because seeds are the main form of planting material for crop production, this technique can be extended to virtually any plant carrying orthodox seeds.
Keywords: Agrobacterium tumefaciens; Cereals; Monocotyledons; Transgenic; Transformation; Wheat.
INTRODUCTION
Cereals represent crops of great importance since they provide more than two-thirds of the world’s food supply (Borlaug, 1998). Demographers forecast that growth of the world population will be lower than expected, but feeding a world of 10 billion people in the 21st century remains a challenge (Borlaug, 2004) since it still requires more calories than the world’s farmers grow today. For this purpose, modern biotechnology must become an integral supplement to conventional breeding programs (Jauhar, 2006). Obtaining viable transgenic cereal plants usable in agriculture is part of the biotechnological effort to ensure food security.
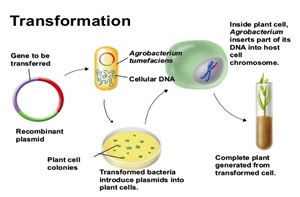
The establishment of useful genetic transformation systems for cereals is of foremost importance for the introduction of traits such as enhanced food value, valorization of low water availability, and increased resistance to disease. Contemporary methods of genetic transformation of cereals, which include the use of attenuated Agrobacterium (Agrobacterium tumefaciens) strains, represent a naturally evolved and artificially optimized vehicle to integrate recombinant DNA into the nuclear genome of plants. Other useful transformation methods include direct gene transfer through the bombardment of target cells with accelerated DNA-coated metal particles.
Agrobacterium-mediated gene transfer into monocotyledonous plants was not possible until recently, since these plants are not among the typical natural hosts of this soil-borne bacterium. Direct DNA-transfer was the preferred principle in most of the early attempts to generate transgenic cereals. In 1994, the pioneering work of Hiei et al. (1994) provided evidence that rice could be efficiently transformed with Agrobacterium and genetic analysis showed the stability of the transformation on R0, R1 and R2 progeny. This finding represented a breakthrough in cereal biotechnology, opening up the possibility of using Agrobacterium for the transformation of major cereal crops such as maize, barley and wheat (For a review, see Shrawat and Lörz, 2006; Goedeke et al., 2007).
Today, the transgenic characteristics of cereals have been reported after transformation through two methods: direct delivery-mediation and Agrobacterium mediation. Both methods can be used to produce transgenic plants with important agronomic traits. Studies on cereal transformation revealed that delivery mediated by particle bombardment facilitated a broad variety of transformation strategies with a wide range of gene expression, and had no biological constraints or host limitations. Various cell types could be targeted efficiently for foreign DNA delivery (Altpeter et al., 2005). Nevertheless, Agrobacterium mediated transformation remains a method of choice for obtaining transgenic plants with a lower copy number, intact foreign genes and stable gene expression.
At this state of advancement in cereal transformation, it seems that Agrobacterium-mediated transformation systems not only promise to intensify attention on the production of cereals with genes of interest, but also to concurrently test and capture the value of the introduced traits (Shrawat and Lörz, 2006). Nevertheless, this transformation method is still cumbersome and time-consuming. The general scheme for Agrobacterium-mediated transformation of cereals plants requires various steps including explants from immature embryos, co-cultivation with the bacteria, selection, regeneration, rooting and transfer to soil. Certain components are necessary for transformation efficiency, such as sugar, plant growth factor, anti-necrotic mixture for controlling oxidative burst, vir-induction chemicals, and antibiotics to suppress or eliminate Agrobacterium following co-culture (for review see Opabode, 2006). At least 13 weeks are necessary from the first explants step to the final planting of a transgenic plantlet in soil.
We describe here an easy and efficient alternative method for obtaining transformed cereal plants based on Agrobacterium-mediated gene transfer, which avoids cell culture and plant regeneration. This is achieved by the incubation of germinating cereal plantlets in a culture of Agrobacterium cells. With this method, we save time, chemicals and media, and the results are similar to those obtained from the classical method.
MATERIALS AND METHODS
Plant material
Seeds from Wheat (Triticum aestivum, cv Soisson), were sterilised by soaking in a 10% sodium hypochlorite for 20 min and washed with sterilised water. They were then incubated in HCl 10 mM solution for 5 min, followed by several washes with sterile distilled water, then germinated on sterile moistened Wattman paper in Petri dish in a chamber at 21 ± 2°C (light/dark).
Isolation of promoter DNA
Sequence promoter of the high molecular weight glutenin subunit (TaHMW) from wheat (cv Soissons) was isolated from genomic DNA by PCR walking adapted from the protocol describing by Devic et al. (1997). Adaptor primers were AP1 5’-GGATCC TAATACGACTCACTATAGGGC-3’; and AP2: 5’-CTATAGGGCTCGAG CGGC -3’. For nested PCR reactions, pfu polymerase (Promega) was used in combination with the specific primers Y1 5’-GATGACTACTGCCGCAAAGAGGAC-3’ and Y2 5’-AAAGAGGACCA GCCGCTTAGCCAT-3’.
The primary PCR parameters were one cycle at 95°C for 3 min, then 30 cycles at 95°C for 1 min, 67°C for 30 sec, 72°C for 1 min, and finally one cycle at 72°C for 7 min and the primers used were AP1 and Y1. For the second PCR, the program was one cycle at 95°C for 3 min, then 30 cycles at 95°C for 1 min, 61°C for 30 sec, 72°C for 1 min, and finally one cycle at 72°C for 7 min and pfu polymerase and the primers used were AP2 and Y2. The fragments obtained were subcloned into the plasmid pGEM-T vector (Promega) for sequencing. Putative cis-regulatory sequences were analyze with bioinformatics approaches identifying transcription factor binding sites or cis-acting sequences in plant promoter using PlantPAN database (http://plantpan.mbc.nctu.edu.tw/index.php, Chang et al., 2008), a system collecting the plant transcription factor binding profiles from PLACE (Higo et al., 1999), TRANSFAC (public release 7.0, Wingender et al., 2000), AGRIS (Davuluri et al., 2003), and JASPER (Sandelin et al., 2004) databases.
GUS reporter gene construction and plant transformation
A 917 pb fragment ending in the ATG codon was amplified by PCR with primers Y3 5’-ATAAAGCTTtggagatcatagaagaacataagag-3’ and Y4 5’- TATGGATC CCATCTCAGTGAACTGTCAGTGA-3’ bearing Hind III and Bamh I restriction sites respectively, and cloned into the pBI101 vector (Clontech) at the initiation codon of the promoterless GUS gene generating the pTaHMWy vector.
For cereal transformation, a procedure was adapted from Clough and Bent (1998). Seeds were allowed to germinate in the dark under controlled, sterile conditions on filter paper soaked with water in Petri dishes. Freshly isolated seedlings were immersed in Agrobacterium suspension for 15 min so that the entire root and shoot systems interact with the Agrobacterium culture, and submit to vacuum to enhance infiltration. Plantlets were washed to remove excess of bacterial solution and transferred on filter paper soaked with water in Petri dishes or into sterilised soil and maintained in a growth chamber at 21 ±2°C under a 14/10 h (light/dark).
Analysis of transgenic plants
Screening of transgenic lines was carrying out by PCR method using a couple of primers created to amplify 491 bp from the GUS gene (5’-GAAAGCGCGTTACAAGAAAGCCG-3’; 5’-GTCGCGCAAGACTGTAACCAC-3’).
Total genomic DNA was isolated from plant leaves using the DNeasy plant Mini Kit (Qiagen). PCR parameters were one cycle at 95°C for 3 min, then 30 cycles at 95°C for 1 min, 61°C for 30 sec, 72°C for 30 sec; and finally, one cycle at 72°C for 7 min.
GUS histochemical staining was performed using 5-brom-4-chloro-3-indolyl-β-D-glucuronide as described by Jefferson et al. (1987). Tissues were then destained with 70% ethanol at 37°C to remove green background. Tissues were observed with Zeiss Axiophot microscope and Leica MZFL III binocular. Images were captured with a Leica DC 200 video camera.
RESULTS
Analysis of TaHMWy promoter
To examine the efficiency of the TaHMW promoter in regulating gene expression, the 5’ flanking sequence was fused to the uidA (GUS) reporter gene via the pBI101 vector and named pTaHMW::GUS (Figure 1).
Figure 1:Schematic representation of the promoter region of the TaHMW gene from wheat (Triticum aestivum, cv Soissons) used in transgenic cereals seedlings. Location of some of the putative cis-acting motifs predicted by PlantPAN database is shown by boxes.
Production of transgenic cereals
The aptitude of cereal seeds for direct transformation was analyzed from the stage of imbibition to that of the developed seedling. Seeds maintained at 4° C were surface sterilized and allowed to germinate under controlled, sterile conditions on filter paper soaked with water. Triticum aestivum cv Soissons, which is responsive to vernalization for flowering, is allowed to germinate at 4°C in the dark. For the transformation procedure, seedlings at different stages of germination were simply immersed in the buffer. Various times of incubation were analyzed between 10 and 45 min., in two cases: under vacuum infiltration, and without it.
We included Silwet L-77 (LEHLE SEEDS), a strong surfactant that shows relatively low toxicity to plants, and often enhances transformation reliability. Plant material was then rinsed with sterile water to eliminate any excess of transformation buffer, and then transplanted in commercial sterile compost for further growth under controlled conditions.
Two weeks after transformation, PCR analysis was performed to screen the different primary plants (T0) grown in commercial sterile compost. A specific 491 bp band corresponding to a GUS sequence was amplified, confirming the integration of the reporter cassette into the recipient genome (Figure 2).
Figure 2:PCR analysis of genomic DNA of T0 progeny by the use of specific primers of GUS gene to amplify a specific 491bp amplicon. T0 transformants were obtained from seedling infected with Agrobacterium tumefaciens strain C58pMP90 (pTaHMW). Plasmid pTaHMW::GUS and genomic DNA from plant transformed with empty cassette were used as positive and negative control respectively. A. Analysis on 1% agarose gels of different lines of transgenic wheat (Triticum aestivum cv Soissons); negative control (1), wheat transformants (2-5), positive control (6), molecular size markers (M).
GUS activity assays in transgenic tissues
Seeds from T0 transgenic plants exhibited various gene reporter expression patterns during germination, according to the plant species. Figure 3 shows a GUS activity in the scutellum and the coleoptile in a germinating wheat seed. In this case, no expression was observed in the endosperm or in radicula.
Figure 3:Transient GUS gene expression in wheat seed from T0 plants expressing the pTaHMW::GUS cassette. A. Negative control. B. GUS activity was visible in florets from young inflorescence. C. GUS Expression observed in the whole immature seed 10 days after anthesis. D. Split seed showing a distribution of blue colour patches in the endosperm in an immature seed (10 days after anthesis).
GUS activity was more visible in florets before anthesis (Figure 3B). It was equally observed in the whole immature seed (Figure 3C). Young seeds cut longitudinally exhibit patches of blue of different intensities (Figure 3C). Analysis of transformed plants by PCR techniques showed that wheat exhibits the greatest aptitude to be transformed by this method with a frequency of 10%.
DISCUSSION
The use of transgenic technologies for the genetic manipulation of plant species has a great impact on biotechnology. Two important components are required for the expression of foreign genes in plant cells: a plasmid carrying an expression cassette and a method to deliver it into the plant cell. Agrobacterium-mediated gene transfer into monocotyledonous plants presents the advantage of singular integration patterns, entailing lower mutational consequences for the transgenic plant and limited transgene silencing. In spite of improvements to render cereal species amenable to genetic engineering, transformation protocols remain tedious.
Seeds are the traditional manner to store cereals and most other plant species, in order to preserve their capacity to germinate. The objective of this study was to develop a rapid method of transformation of cereals starting from seed, thus avoiding the stages requiring in vitro tissue culture and plant regeneration. The transformation events reported in this work produced fully fertile plants expressing the reporter gene and transmitting it to T1 progeny, where it was also expressed. Furthermore, the transformation efficiency is similar to what is generally found with other methods for wheat and maize, and even better for sorghum (Sahrawat and Lorz 2006; Nguyen et al., 2008).
Transformation frequency observed by the direct method showed that it was relatively easy to transform the cereals varieties used. Surfactants may enhance T-DNA delivery by aiding Agrobacterium attachment and or by elimination of certain substances that inhibit bacteria attachment (Opabode, 2006). These results constitute a good argument for the method, because to achieve transgenic lines remains in itself a challenging task, due to the low transformation frequency (Cheng et al., 2004).
CONCLUSION
In summary, the technology for genetic engineering of plants has an important impact on basic research and biotechnology. The transformation procedure for plant models, such as Arabidopsis, is under control today. Achieving easy and effective methods of transformation for important plant crops, which are often the most recalcitrant species, is necessary for their study and their improvement. In this context, the techniques of transformation of cereals constitute a very active research field, to develop versatile binary vectors and new explants, or to take advantage of specific regulating elements found in promoters driving gene expression in seed tissues. In this paper, we developed a procedure allowing the direct transformation of cereals by Agrobacterium tumefaciens. This method avoids the tissue culture and plant regeneration steps, constituting time savings of at least ten weeks, compared with the requirements of the standard method. Furthermore, it saves reagents and handling. The method is easy and effective for the expression of GUS gene in wheat. This method of transformation is potentially applicable to any plants species carrying orthodox seeds.
REFERENCES
Altpeter F., Baisakh N., Beachy R., Bock R., Capell T., Christou P., ... & Fauquet C. (2005). Particle bombardment and the genetic enhancement of crops: myths and realities. Molecular Breeding, 15: 305-327.
Borlaug N.E. (1998). Feeding a world of 10 billion people: the miracle ahead. Plant Tiss. Cult. Biotech., 3: 119–127.
Borlaug N.E. (2004). Feeding a world of 10 billion people: our 21st century challenge. Perspectives in world food and agriculture, 31-56.
Cheng M., Lowe B.A., Spencer T.M., Ye X., Armstrong C. L. (2004). Factors influencing Agrobacterium-mediated transformation of monocotyledonous species. In Vitro Cellular & Developmental Biology-Plant, 40: 31-45.
Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. The plant journal, 16: 735-743.
Devic M., Albert S., Delseny M., Roscoe T.J. (1997). Efficient PCR walking on plant genomic DNA. Plant physiology and biochemistry (Paris), 35: 331-339.
Goedeke S., Hensel G., Kapusi E., Gahrtz M., Kumlehn J. (2007). Transgenic barley in fundamental research and biotechnology. Transgenic Plant J., 1: 104-117.
Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. The Plant Journal, 6: 271-282.
Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic acids research, 27: 297-300.
Jauhar P. P. (2006). Modern biotechnology as an integral supplement to conventional plant breeding: the prospects and challenges. Crop Science, 46: 1841-1859.
Jefferson R.A., Kavanagh T.A., Bevan, M.W. (1987). GUS fusions: beta‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO journal, 6: 3901-3907.
Nguyen T.V., Thu T.T., Claeys M., Angenon G. (2007). Agrobacterium-mediated transformation of sorghum (Sorghum bicolor (L.) Moench) using an improved in vitro regeneration system. Plant cell, tissue and organ culture, 91: 155-164.
Opabode J.T. (2006). Agrobacterium-mediated transformation of plants: emerging factors that influence efficiency. Biotechnol. Mol. Biol. Rev., 1: 12-20.
Sandelin A., Alkema W., Engström P., Wasserman W. W., Lenhard, B. (2004). JASPAR: an open‐access database for eukaryotic transcription factor binding profiles. Nucleic acids research, 32: D91-D94.
Shrawat A.K., Lörz H. (2006). Agrobacterium‐mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnology Journal, 4: 575-603.